Epydemix
The ABC of Epidemics
Epydemix provides a unified, open-source framework that enables researchers and public health practitioners to design, simulate, and calibrate epidemic models within a single environment.
Python Package
epydemix
An open-source Python package designed for flexible, modular, and data-driven epidemic modeling.
Epydemix supports the full modeling pipeline—from constructing stochastic compartmental models to running simulations, integrating real-world data, and calibrating parameters. Users can incorporate age-structured contact patterns, dynamic interventions, and population demographics with ease. Built-in Approximate Bayesian Computation (ABC) methods enable robust parameter estimation and model fitting, supporting forecasting, scenario exploration, and policy-relevant analyses. Epydemix bridges the gap between theoretical modeling and practical application, helping researchers and public health professionals translate models into actionable insights.
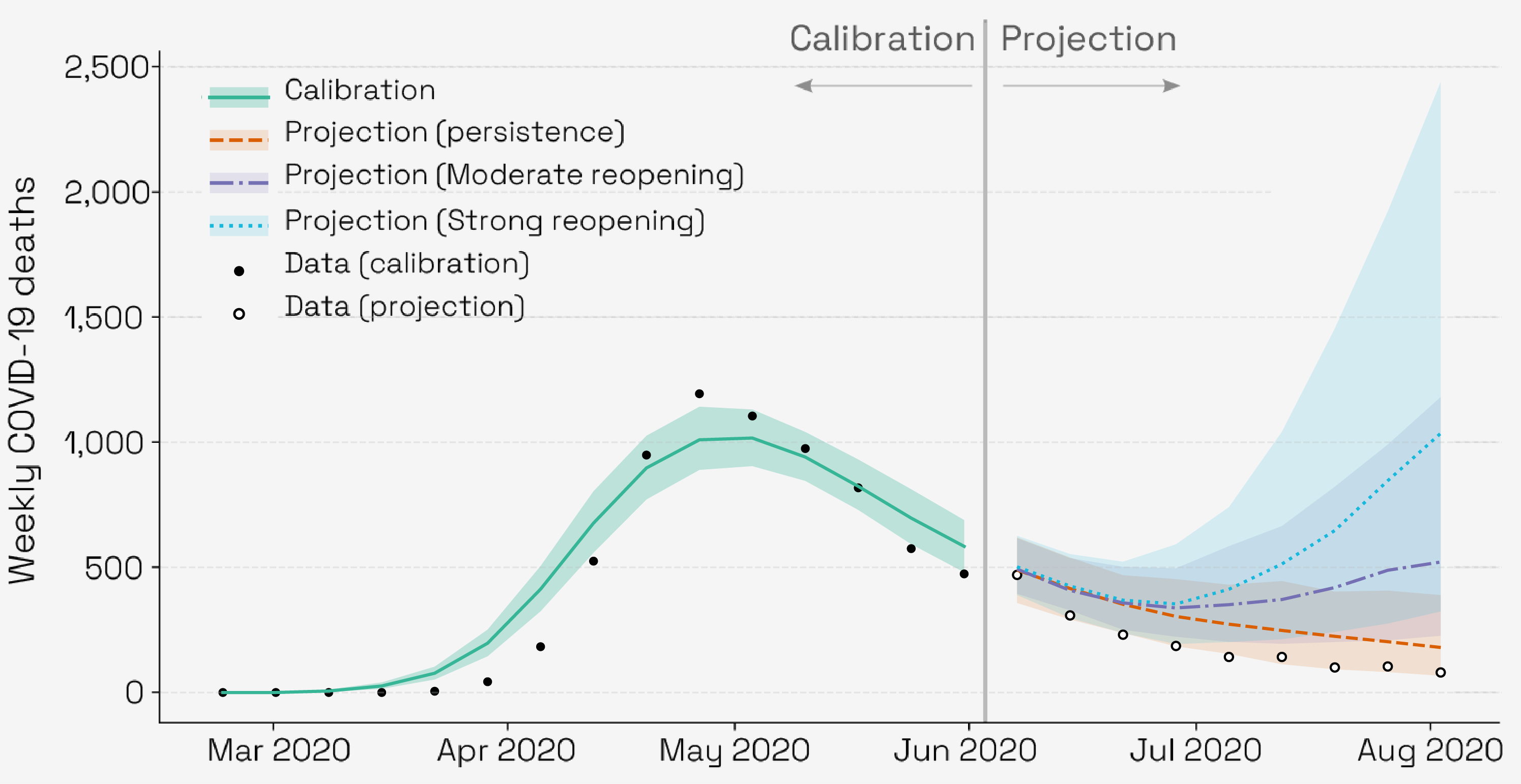
Data
Epydemix includes access to population data and synthetic age-stratified contact matrices for over 400 countries and regions worldwide. These datasets enable users to construct realistic, demographically grounded epidemic models. Contact matrices capture interactions across key settings — home, work, school, and community — and are provided in multiple formats to support a range of modeling scenarios.
Papers & scientific outputs
Nicolò Gozzi, Matteo Chinazzi, Jessica T. Davis, Corrado Gioannini, Luca Rossi, Marco Ajelli, Nicola Perra, Alessandro Vespignani
PLOS Computational Biology 21, no. 11 (2025): e1013735. view pdf




